Properties: Charge Density (ECH3)¶
The CryEch3Calculation can be used to run the properties
executable for ECH3 calculations, from an existing fort.9.
See also
Properties Workflow to run multiple properties calculations (and optional initial SCF).
!verdi plugin list aiida.calculations crystal17.ech3
Inputs
code: required Code The Code to use for this job.
parameters: required Dict the input parameters to create the properties input file.
wf_folder: required FolderData, RemoteData, SinglefileData the folder containing the wavefunction fort.9 file
metadata: optional
Outputs
charge: required GaussianCube The charge density cube
remote_folder: required RemoteData Input files necessary to run the process will be stored in this folder node ...
results: required Dict Summary Data extracted from the output file(s)
retrieved: required FolderData Files that are retrieved by the daemon will be stored in this node. By defa ...
spin: optional GaussianCube The spin density cube
Exit codes
1: The process has failed with an unspecified error.
2: The process failed with legacy failure mode.
10: The process returned an invalid output.
11: The process did not register a required output.
200: The retrieved folder data node could not be accessed.
210: The main (stdout) output file was not found
211: The temporary retrieved folder was not found
300: An error was flagged trying to parse the crystal exec stdout file
350: The input file could not be read by crystal
351: Crystal could not find the required wavefunction file
352: Parser could not find the output density file
353: Error parsing output density file
400: The calculation stopped prematurely because it ran out of walltime.
401: The calculation stopped prematurely because it ran out of memory.
402: The calculation stopped prematurely because it ran out of virtual memory.
413: An error encountered usually during geometry optimisation
414: An error was encountered during an scf computation
415: An unknown error was encountered, causing the mpi to abort
499: The main crystal output file flagged an unhandled error
from aiida import engine, load_profile, orm, plugins
from aiida.tools.visualization import Graph
from aiida_crystal17.common import recursive_round
from aiida_crystal17.tests.utils import (
get_or_create_local_computer, get_or_create_code,
get_default_metadata)
from aiida_crystal17.tests import open_resource_binary
profile = load_profile()
profile.name
'test_crystal17'
Running a calculation¶
The ECH3 Input Schema gives the allowed format of the input dictionary, for example:
computer = get_or_create_local_computer('work_directory', 'localhost')
code = get_or_create_code('crystal17.ech3', computer, 'mock_properties17')
builder = code.get_builder()
builder.metadata = get_default_metadata()
builder.parameters = orm.Dict(dict={
'npoints': 20
})
with open_resource_binary('ech3', 'mgo_sto3g_scf', 'fort.9') as handle:
builder.wf_folder = orm.SinglefileData(handle)
result, calcnode = engine.run_get_node(builder)
!verdi process show {calcnode.pk}
Property Value
------------- ------------------------------------
type CalcJobNode
pk 974
uuid 4dd70b8f-e7b6-4684-84e2-69f5db4ddbb8
label
description
ctime 2019-09-24 15:45:47.628170+00:00
mtime 2019-09-24 15:46:01.609931+00:00
process state Finished
exit status 0
computer [1] localhost
Inputs PK Type
---------- ---- --------------
code 971 Code
parameters 972 Dict
wf_folder 973 SinglefileData
Outputs PK Type
------------- ---- ------------
charge 978 GaussianCube
remote_folder 975 RemoteData
results 977 Dict
retrieved 976 FolderData
graph = Graph(graph_attr={'size': "6,8!", "rankdir": "LR"})
graph.add_node(calcnode)
graph.add_incoming(calcnode, annotate_links="both")
graph.add_outgoing(calcnode, annotate_links="both")
graph.graphviz
Analysing the outputs¶
The outputs are:
results a dict of computation input and output parameters, parsed from the stdout file.
charge a data node containing the gaussian cube file for the charge density.
spin a data node containing the gaussian cube file for the spin density (if the original computation included spin).
recursive_round(calcnode.outputs.results.get_dict(), 1)
{'units': {'energy': 'eV', 'conversion': 'CODATA2014'},
'errors': [],
'header': {'crystal_version': 17, 'crystal_subversion': '1.0.1'},
'warnings': [],
'wf_input': {'n_ao': 14,
'n_atoms': 2,
'k_points': [8, 8, 8],
'n_shells': 5,
'n_symops': 48,
'gilat_net': 8,
'n_core_el': 12,
'n_electrons': 20,
'energy_fermi': -4.0,
'energy_total': -7380.2,
'n_kpoints_ibz': 29,
'energy_kinetic': 7269.0},
'parser_class': 'CryEch3Parser',
'parser_errors': [],
'parser_version': '0.11.0',
'parser_exceptions': [],
'execution_time_seconds': 0}
The GaussianCube data node stores a gaussian cube in a compressed zip file.
calcnode.outputs.charge.attributes
{'cell': [[0.0, 2.2157919831613, 2.2157919831613],
[2.2157919831613, 0.0, 2.2157919831613],
[2.2157919831613, 2.2157919831613, 0.0]],
'units': {'length': 'angstrom', 'conversion': 'CODATA2014'},
'header': ['Charge density - 3D GRID - GAUSSIAN CUBE FORMAT MgO Bulk',
'5.62556267 5.62556267 5.62556267 60.000000 60.000000 60.000000'],
'elements': ['Mg', 'O'],
'voxel_grid': [20, 20, 20],
'zip_filename': 'gcube.zip',
'cube_filename': 'gaussian.cube',
'compression_method': 8}
The full file can be accessed via:
with calcnode.outputs.charge.open_cube_file() as handle:
print(handle.readline())
Charge density - 3D GRID - GAUSSIAN CUBE FORMAT MgO Bulk
There is also methods available to parse the file to a dict or structure:
data = calcnode.outputs.charge.get_cube_data()
data.atoms_atomic_number
[12, 8]
calcnode.outputs.charge.get_ase()
Atoms(symbols='MgO', pbc=True, cell=[[0.0, 2.2157919831613384, 2.2157919831613384], [2.2157919831613384, 0.0, 2.2157919831613384], [2.2157919831613384, 2.2157919831613384, 0.0]])
Some experimental methods also exist, for basic analysis of the density.
calcnode.outputs.charge.compute_integration_cell()
18.60283258916424
calcnode.outputs.charge.compute_integration_atom([0, 1], radius=1.0)
[17.16956646407823, 1.1837163766941237]
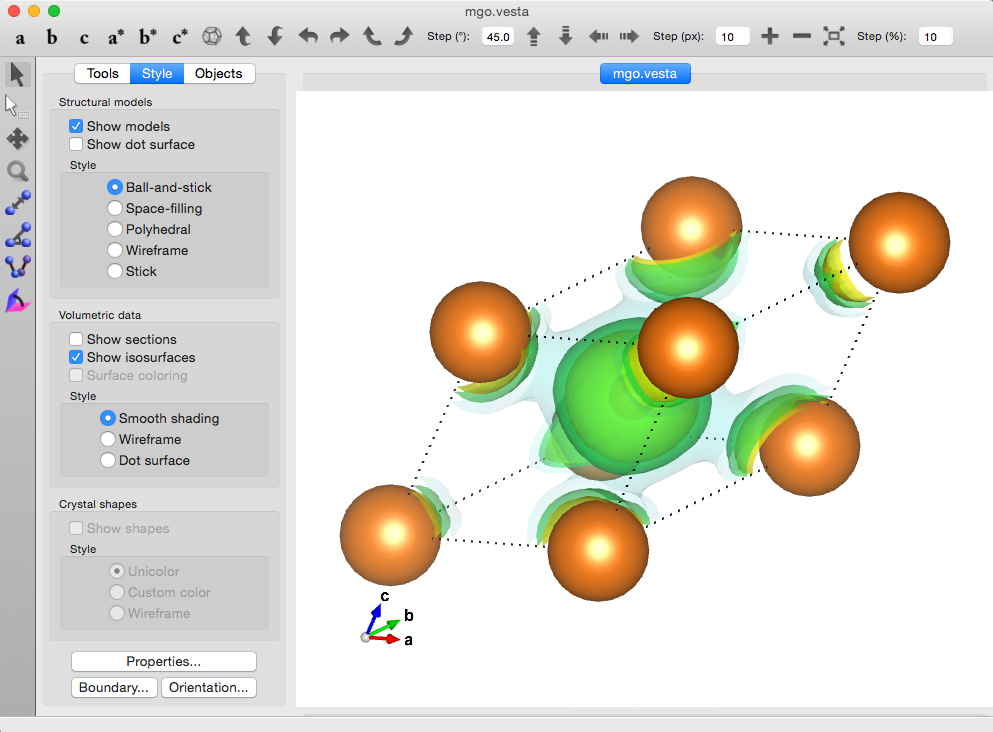
Visualising the density in VESTA¶
The vesta module
contains functions, to convert cube data to input files that can be opened in VESTA.
The VESTA Settings Input Schema gives the allowed format of the input settings dictionary, for example:
from aiida.common.folders import SandboxFolder
from aiida_crystal17.parsers.raw.vesta import (
create_vesta_input, write_gcube_to_vesta)
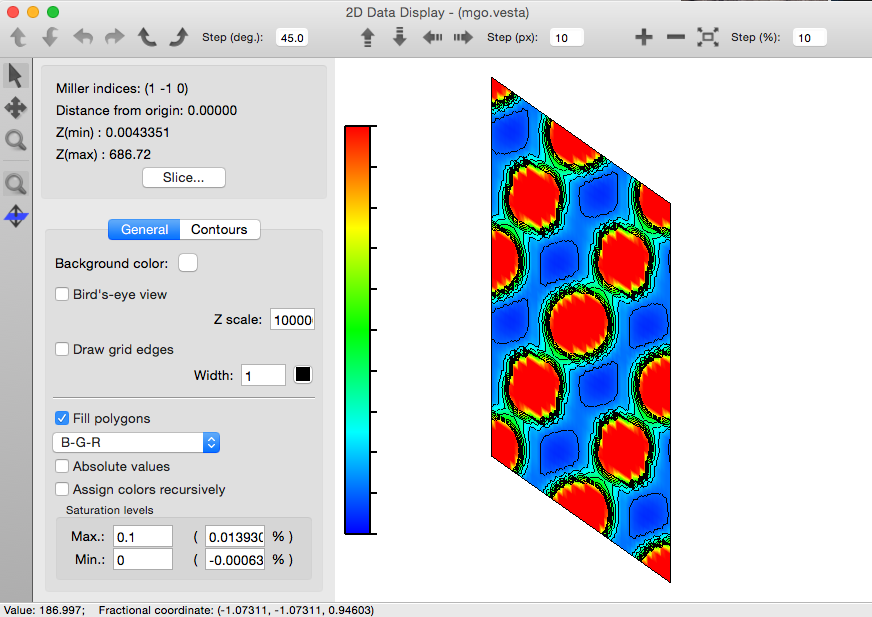
settings = {
"2d_display": {
"h": 1.0,
"k": -1.0,
"l": 0.0,
"dist_from_o": 0.0
}
}
with SandboxFolder() as folder:
write_gcube_to_vesta(
calcnode.outputs.charge,
folder.abspath, 'mgo', settings)
print(folder.get_content_list())
with folder.open('mgo.vesta') as handle:
print(handle.read()[:100])
['mgo.cube', 'mgo.vesta']
#VESTA_FORMAT_VERSION 3.3.0
CRYSTAL
TITLE
GAUSSIAN_CUBE_DATA
IMPORT_DENSITY 1
+1.000000 mgo.cube